Seasonal Variation of Dolphinfish Stocks (Coryphaena hippurus) in the Pacific Coast of Colombia- Juniper Publishers
Juniper Publishers- Journal of Oceanography
Abstract
Large pelagic fish are characterized by large population sizes, high dispersal capabilities and cosmopolitan distributions, decreasing the chances for population structuring. Therefore, considering ecological factors that can influence gene flow is important to understand possible stock structure. For the dolphinfish (Coryphaena hippurus) migratory movements attributed to sea changing conditions have been documented in the Eastern Caribbean. In the Colombian Pacific there has been no evaluation of possible stock differentiation. In order to adequately answer this question, temporal scale must be taken into consideration since seasonal abundance of this fish occurs in the Pacific coast of Central and South America. Here we investigate possible seasonal variation of dolphinfish stocks via analyses of mitochondrial DNA sequence data and analyses of five microsatellite loci. A total of 128 specimens were sampled from the coast of Colombia between November 2010 and December 2011. Levels of genetic differentiation between all sampling dates (months) were analyzed by calculating pairwise FST in order to detect genetic heterogeneity in a temporal scale. These analyses suggested subtle population differentiation among samples obtained in the first months of the year (Nov-May) vs. samples from months in the mid part of the year (Jun-Oct). Additionally, the Bayesian assignment test showed that the higher value of K was for K=2. Both analyses showed genetic heterogeneity in a temporal scale, suggesting the presence of possibly two different stocks in the Pacific Coast of Colombia along the year. As our data suggests there is an incursion of individuals into the Pacific of Colombia during the seasonal abundance peak of capture between the months of January and May. These results suggest that individuals collected in close dates of the year are significantly less different among them than individuals collected in dates far apart. Considering dolphinfish are moving in the Pacific and each country has not only different management strategies but a different total allowable catch, the combined numbers of total allowable catch may end reducing the entire fish population and threatening the stock size if the information of possible stock differentiation is not considered in local and regional management plans.
Keywords: Dolphinfish; Coryphaena hippurus; Fish stocks; Pacific ocean; Microsatellites; Mitochondrial DNA
Introduction
Fish, like most marine resources, are increasingly growing to depletion due to intensified overfishing [1]. Therefore, it has becomes necessary to define the most appropriate way to manage fisheries in a sustainable way using both genetic and ecological approaches. In this sense, management of wild fisheries should be considered as a main priority in order to ensure that local fisheries can continue to be exploited in the long run [2]. Moreover, if research is conducted to identify the species genetic and ecological characteristics, this information could then be used on the recovery of depleted stocks and management of healthy ones [3].
Biological stocks are defined as "an intraspecific group of randomly mating individuals with temporal and spatial integrity" [4]. However, in most cases, fishery managers tend to define stocks as fish co-occurring in the same fishing ground, without taking into consideration their genetic integrity. Identifying stocks as not simply co-occurring individuals in a given area at a specified time but as individuals sharing similar genetic and ecological characteristics provides insight into more effective management due to true biological stock assessment [5].
Genetic and ecological analyses have been useful as an unequivocal and objective tool for assessment of true biological stocks. This has been used recently in stock definition in some large, pelagic, migratory and widely distributed fishes such as the striped marlin ((kajikia audax), blue marlin (Makaira nigricans) and bluefin tuna (Thunnus thynnus) [6,7]. The biological attributes of these species include:
- They form large spawning congregations,
- There is absence of apparent barriers to gene flow and
- They have an intrinsic ability for large-scale dispersal.
From these characteristics, one would expect a lack of genetic structure in large marine pelagic fish including the later species. However, in a number of these species genetic studies have shown the existence of more than one genetic biological stock [6,8,9]. For example, in the bluefin tuna, Appleyard et al. [8] showed the existence of significant spatial heterogeneity within the Mediterranean Sea. Further research into these species, that have been assumed to form large panmictic populations and are the main objective for fisheries around the world, should be considered as a priority to define stocks to be used in fisheries management programs.
The dolphinfish (Coryphaena hippurus), also known as "dorado" or "mahi-mahi", is a cosmopolitan, migratory pelagic fish found in tropical and subtropical waters [10]. In the Northeast Pacific Ocean this species is distributed throughout the tropical region from the Equator to Baja California. As other such large pelagic fish, dolphinfish exhibits high growth rates, early maturity, short life span and capacity for releasing several successive batches of eggs [11]. These features should, in theory, reduce the possibility of developing genetic differences between populations. However, conflicting data suggest the existence of regional factors that may limit connectivity and promote genetic heterogeneity as has been previously found in studies on dolphinfish in the Eastern Tropical Pacific and Gulf of California [12,13] or this reason, it is then important to correctly assess whether the Pacific population consists of a single panmictic population or if several discrete populations can coexist at local scales.
It has been demonstrated that dolphinfish exhibit seasonal abundance in different countries, which has been explained by a possible migratory movement along the Eastern Pacific Coast[14] .Migratory routes such as the one suggested for the Eastern Pacific Coast have been already proved in the Caribbean. Farrel[15] found structured movement patterns inferred from tagged individuals and by analyses of ecological niche modeling.
The seasonal abundance observed in different countries may be the result from movement patterns of dolphinfish in the Pacific. This seasonal abundance has been seen in countries like Costa Rica (Sept-February) and Ecuador (November-May). In Colombia the biggest peak in catch rate is between the months of January and April; yet dolphinfish individuals captures show higher catch rates starting in November. Dolphinfish are captured throughout the year, which implies there must be a local resident group in the Pacific Coast of Colombia [14]. Even if global studies on dolphinfish show lack of interoceanic differentiation [16], this does not necessarily imply that there are not locally and discrete populations along the Pacific Coast. In fact, a monthly population assessment may increase the potential for finding stock structure if the species exhibits temporally and spatially constrained demographic pulses [12].
Dolphinfish is considered an important fishery resource on the Pacific Coast of Colombia, being one of the dominant fish obtained from the artisanal and industrial fleet landings. It is considered the second most important resource on the Pacific Coast of Colombia after the snapper (Lutjanidae), and above sharks (Carcharhinidae). Although its annual catch contribution is less than that of tuna, its annual landings are between 300 and 600 tons, making it a valuable resource in Colombia [14]. Based on the seasonal movement patterns presented by dolphinfish in the Pacific Coast of Colombia and the genetic data obtained in this study, we were able to test two hypothesis regarding dolphinfish stock structure in the Pacific Coast of Colombia. These were:
- Dolphinfish form relatively discrete populations (stocks) in this area or
- They all belong to a single panmictic population in the Pacific Coastal waters of Colombia where the fishery takes place.
In the specific case of dolphin fish, multiple fisheries compete to catch fish in different countries; therefore understanding the behavior of exploited fisheries is key in order to implement management strategies. In order to test these two hypotheses, we analyzed five microsatellite loci and two mitochondrial genes (NADH1 dehydrogenase subunit 1 (ND1) and cytochrome b (cytb)) for 117 dolphinfish captured between November 2010 and December 2011, along the Pacific Coast of Colombia.
Materials and Methods
Sample collection
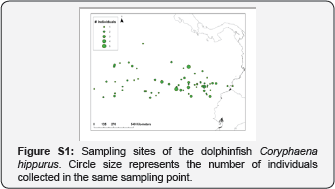
Samples of dolphinfish were collected in the Pacific Coast of Colombia over one consecutive year (2011) from small fishing boats operating in a local scale in the Pacific Coast of Colombia. Observers from the Instituto Colombiano de Desarrollo Regional (INCODER) collected muscle tissue samples (2g) from 128 dolphinfish between November 2010 and December 2011 (Supplementary material (SM); Figure S1). Tissue samples were preserved in 80�x0025; ethanol until DNA extraction. Accompanying data of the samples included GPS coordinates of the capture location, sex and size (cm) of the animal (Table S1).
DNA data collection
Mitochondrial gene regions: Genomic DNA was extracted following a phenol-chloroform protocol [17]. A 751 base pairs (bp) fragment of the NADH1 dehydrogenase subunit 1 (ND1) gene of dolphinfish was amplified using the primers NADH163 (5'-TAATCCTGCCGCAATTATCC-3') and NADH1128 (5'AGGCCTTCCAGGTTAGGT GT-3') designed by Diaz et al. [16]. PCR reactions were made up to a total volume of 30ml containing 10-100ng DNA. PCR amplification conditions consisted of 35 cycles of 1min at 95 °C for denaturation, 1min at 58.9 °C for annealing and a final extension at 74 °C for 3mins. A 500 base pair (bp) fragment of the cytochrome b (cytb) gene of dolphinfish was amplified using the internal primers (L14841 and H15149) [18]. Amplification reactions were made up to a total volume of 30ml containing 10-100ng of DNA. Amplification conditions consisted of 35 cycles of 2min at 94 °C for denaturation, 45sec at 55 °C for annealing and a final extension at 72 °C for 10mins. Amplicons were purified with polyethylenglycol and sequenced on an ABI 3770 Xl automated sequencer (Applied Biosystems) by Macrogen Inc., Korea.
Microsatellite genotyping: Five dolphinfish microsatellite loci registered in GenBank (Robert Chapman; South Carolina Department of Natural Resources, Charleston SC., unpublished) (Chi002, Chi008, Chi008a, Chi023 and Chi037) already used in dolphinfish [13] where amplified. Amplifications took place in 12, 5ml reactions with the following concentrations: 1x buffer, 3mm MgCl2, 00.2mm dNTP, 0.003mm of each primer and 0,002ul-1 Taq polymerase. The cycling profile consisted of an initial denaturing period of 2min at 94 °C, five cycles at 94 °C for 5s, annealing for 60s at 59 °C and extension for 60s at 72 °C, 20 cycles at 94 °C for 0s, annealing for 60s at 59 °C and extension for 60s at 72 °C, and a final extension for 60min at 59 °C. Products were run on an ABI3100 sequencer at Universidad de los Andes. Allele size was measured against an internal ladder (Tamra 500) using Gene Scan Analysis Software (1998). The binning of the microsatellite data was performed using the software TANDEM that uses a heuristic search with the Nelder-Mead Downhill Simplex algorithm and applies a least-square minimization of rounding errors to determine the allele number [19]. The program MICRO-CHECKER v. 2.2.3 [20] was used to determine the presence of null alleles, large allele dropout and scoring errors due to stutter peaks.
Data Analysis
Mitochondrial DNA
Sequences were aligned using Geneious Pro 3.6.1 [21] and edited manually. Unique haplotypes and variable sites were identified using MacClade v. 4.0 [22]. Haplotype (h), and nucleotide (n) diversities were estimated with Arlequin 3.0 [23]. Hierarchical genetic structure of the samples was undertaken by assessing the relative contribution among groups, within groups, and within populations (AMOVA) using Arlequin 3.0 [23]. For this, we specifically tested the hypothesis based on the seasonal abundance peak, which considers individuals sampled between Dec and April (seasonal abundance peak) vs individuals sampled between May and Oct. 10,000 permutations.
Microsatellite loci
Levels of genetic diversity, expressed in number of alleles (NA), observed heterozygosity (H0) and expected heterozygosity (HE) were obtained for each locus. Testing for deviations from Hardy-Weinberg equilibrium was performed using the software Arlequin 3.0 [23]. The null hypothesis of independence between loci (no linkage disequilibrium) was tested using the same software.
Levels of genetic differentiation between all sampling dates (months) were analyzed by calculating Wrigth's pairwise FST and the D estimate (actual differentiation of Jost 2008) was calculated with SMOGD [24]. A global AMOVA test including all samples was conducted to obtain the global FST value. Hierarchical genetic structure of the samples was undertaken by assessing the relative contribution among groups, within groups, and within populations (AMOVA) using Arlequin 3.0 [23]. For this, we specifically tested the hypothesis based on the seasonal abundance peak, which considers individuals sampled between Dec and April (seasonal abundance peak) vs individuals sampled between May and Oct. 10,000 permutations.
A Bayesian approach for assignment of individuals into stock was performed using the software Structure 2.3.1 [5,7,25] to complement the results obtained with F statistics. The admixture model and correlated allele frequencies between populations was selected. The Markov Chain Montecarlo (MCMC) consisted of 100000 steps with a burn-in period of 25,000 steps. We explored a range of K from one to seven with six runs for each K-value. The appropriate number of clusters (K) was selected using the criterion of Evanno [26] using the STRUCTURE HARVESTER v0.56.3 software [27].
Results
Mitochondrial DNA
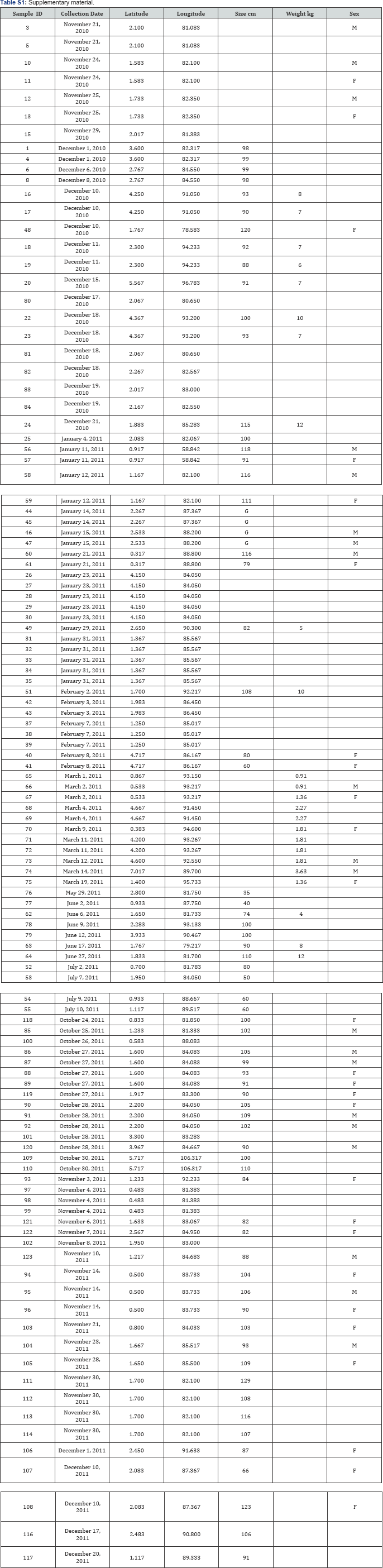
Genetic diversity: A 525bp segment of ND1 gene was sequenced for 67 dolphinfish. A total of 61 variable sites defined 33 haplotypes. Haplotype diversity averaged h = 0.9109, and nucleotide diversity averaged n=1.31 %. For cytb a 345 base pair segment was sequenced for 78 dolphinfish. A total of 131 variable sites defined 35 haplotypes. Haplotype diversity averaged h=0.9108 and nucleotide diversity averaged n=1.87 %. Haplotype frequencies per month for cytb (Table 1) and ND1 gene (Table 2) showed that between the months of December to February, during the seasonal abundance peak in Colombia [14], a higher total number of haplotypes was found. For cytb the most frequent haplotypes during these months were dorA, dorB, dorC and dorD. From these, only haplotypes dorA, dorB and dorC were also found in samples collected in October, November and December 2011 when the seasonal abundance peak started again. Three unique haplotypes were reported for June and July, months in which the catch rate is severely reduced. These unique haplotypes were not present at any other time during the year. For ND1 the most frequent haplotypes between the seasonal abundance months were dorA and dorD. Together with dorB, C, D, E, F and G they were only present between the months of December to March. Two unique haplotypes were reported as well for Jun and July. ND1 and cytb haplotypes defined in this study were submitted to Genbank as accession numbers XX to YY.

Additional haplotypes represented by only one individual per month; for 2010 (Nov:1, Dec:7), for 2011 (Jan:6, Feb:2, Mar:3, Jun:2, Jul:1).
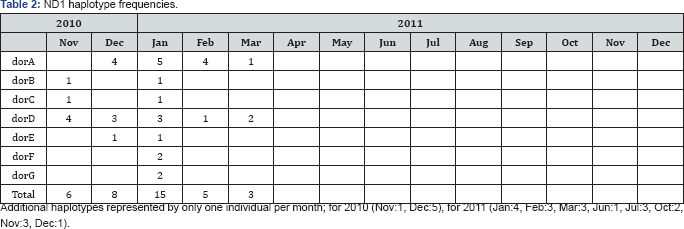
Genetic differentiation: Pairwaise FST comparison between sampled dates (months) for ND1 gene (Supplementary Material, SM; Table S2) showed small but significant differences between November 2010 vs March 2011, December 2010 vs June 2011 and June 2011 vs November and December 2011. These differences are mainly between those individuals captured during the high catch season (January-April) and the ones captured in the other months. Most of the total genetic variance was found within populations (FST=0.042, P=0.049). The hypothesis that genetic variation could be structured according to the seasonal abundance peak was not rejected (FCT=0.124, P=0.032). For the cytb gene pairwaise FST comparisons (SM; Table S3) showed small and significant differences between November 2010 vs December 2010, December 2010 vs January 2011, and November 2010 vs May 2011. For cytb gene the hypothesis considering the genetic variation could be structured according to the seasonal abundance peak was rejected.
Numbers in bold correspond to significant FST values (p<0.002). Underlined Nov and Dec correspond to 2010.
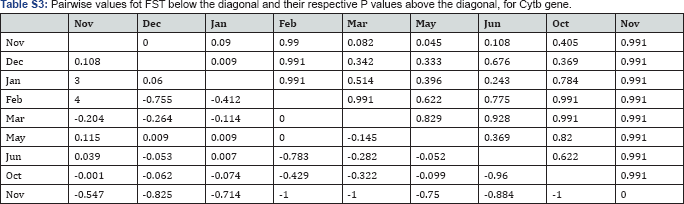
Numbers in bold correspond to significant FST Values (p<0.005).
Microsatellites
Results from micro-checker showed that null alleles were not present. The analysis showed no evidence or large allelic dropout or stuttering. On average, the observed heterozygosityz (HO) was lower than the expected heterozygosity (HE). The observed heterozygosity (HO) ranged from 0.85 to 1 and expected heterozygosity (HE) ranged from 0.85 to 0.94 (Table 3). The number of alleles at each locus ranged from fifteen to forty-two. No loci were found to be in linkage disequilibrium (P>0.05) and all five loci appeared to deviate from Hardy- Weinberg equilibrium. Only two loci (chi008 and chi023) were in HWE equilibrium after Bonferroni's correction (adjusted P value = 0.01).

Sample Size (N): Number of Alleles; (NA): Observed Heterozygosity; (H0): Expected Heterozygosity; (HE): Hardy Weinberg Disequilibrium was observed for the five loci used in this study (P>0.05).
Pairwise FST comparison between sampled dates (months) for microsatellites (SM; Table S4), showed significant differences between fish caught in Dec2010, January, February, March and July versus fish caught in November. Fish caught in November are still considered to be part of that time of the year where the seasonal abundance peak has not begun. Likewise between February and October significant differences where observed. All these estimates were consistent with the D estimator. The AMOVA analysis revealed that most of the total genetic variance was found within populations (months) (FST=0.011,P=0.0009) therefore the hypothesis that nuclear variation could rejected (FCT=-0.0024, P=0.00587). Such negative estimates should be interpreted as zero in the AMOVA [23]. Surprisingly be structured according to the seasonal abundance pattern was July appeared to be significantly different from all other months.

Numbers in bold correspond to significant FST values (p<0.05).
The assignment test analysis conducted in STRUCTURE for the Colombian samples showed the highest likelihood at a population structure of K=2 which was then confirmed using the Evanno et al. [26] method. The peak in the distribution of K was found at K=2 (Figure 2).

Discussion
The mean time of onset of sensory blockade and motor blockade between the two groups is comparable with p value of (0.3843) and (p=0.2133) which is not statistically significant. Two segment regression of sensory blockade is significantly prolonged by addition of intrathecalnalbuphine as seen in group N. The duration of analgesia is significantly prolonged with addition of nalbuphine compared with bupivacaine alone.The degree of population differentiation has been related to the dispersal capacity of the species [28]. It has been recognized that species with higher dispersal capabilities show reduced levels of population differentiation [29]. This pattern has been observed in other large pelagic fishes such as tuna and billfishes [6,8]. Dolphinfish populations have been characterized by having great dispersal capabilities and effective population sizes similar to those observed in other pelagic fishes, as those estimated for bigeye tuna [30]. Large effective population sizes and great dispersal capabilities are biological attributes that may explain the reduced observed levels of population differentiation in dolphinfish [16]. The approach used in this study, taking into consideration the possible migratory capabilities observed in dolphinfish, has provided new insights into how dolphinfish populations are temporarily structured at least for populations found in the Pacific Coast of Colombia.
In this study we provide information about the genetic diversity and population structure of dolphinfish C. hippurus in the Pacific Coast of Colombia. Our analysis supports the hypothesis of the Pacific Coast of Colombia having two discrete temporally spaced stocks of dolphinfish, although at this time we have no evidence to define if individuals collected in distant months of the year belong to reproductively isolated units.In this study we found high heterozygosity in the five loci analyzed (HE = 0.77), this high levels of genetic variation have also been observed in dolphinfish [13] and in other large pelagic fish as big eye tuna [30]. The deviation towards heterozygous excess may be a result of mixing of two discrete isolated stocks, causing a temporary excess of heterozygotes. However, our data shows a clear tendency towards hardy-weinberg disequilibrium, perhaps as a result from the incursion of individuals from distant places, having a strong effect on the genetic composition of dolphinfish catches along the year in the Pacific Colombian Coast.
Both mitochondrial genes ND1 and cytb revealed a higher number of haplotypes during the seasonal abundance peak. This is in agreement with the higher number of individuals present during these months of the year, as has been suggested for dolphinfish catch records in the Pacific of Colombia [14]. Particularly for cytb, the most frequent haplotypes during the catch peak between the months of January and April (dorA, dorB and dor C) were also present at Oct, Nov and December from 2011 when the seasonal abundance peak started again. This corroborates that the seasonal abundance peak starts in late November an lasts until April, as evidenced in catch records for dolphinfish [14]. For both ND1 and cytb unique haplotypes were present along the sampling period. Interestingly, there were also unique haplotypes for months when the catch rate is reduced. This would be suggestive of shared population history between stocks in the Eastern Pacific.
The change in allelic frequencies along the sampling year can be observed with the pairwise distance FST and the bayesian assignment test. Both analyses showed slight genetic heterogeneity in a temporal scale, suggesting the presence of possibly two different stocks in the Pacific Coast of Colombia along the year. Our results support there is a considerable change in allelic frequencies along the year, with low but significant (FST = 0.011, P =0.0009) values. As our data suggest there is an incursion of individuals into Colombian territory, during the seasonal abundance peak between the months of January and May [14]. This seasonal abundance in the Colombian Coast seems to be due to the arrival of dolphinfish individuals with different allelic frequencies as inferred with the Bayesian Assignment tests. Additional data from other Central (Mexico, Costa Rica and Panama) and South American countries (i.e. Ecuador) is necessary in order to infer and confirm the movement patterns of dolphinfish in the Pacific.
It has been demonstrated that dolphinfish move long distances in the Caribbean, as a result to changing sea surface temperature (STT). Moving southward during the colder months (August to February) and moving backwards to the Coast of Florida during the warmer months (May to July) [15]. In the Pacific Coast of Colombia dolphinfish possible migrations have been associated with thermal fronts forming in the Colombian Coast along the year. Also, the most persistent thermal fronts, where higher catch rate of dolphinfish have been reported, are more intense during the first two trimesters of the year (December-February and March-May) [31]. These months coincide with the peak of seasonal abundance of dolphinfish in the Pacific coast of Colombia. Considering this information, it is then plausible that dolphinfish stock dynamics in the Pacific are affected by similar factors such as the ones modeling the migration movements of this species in the Caribbean. Further research including satellite tagging would provide better understanding into the biology of this species in the Colombian Pacific.
A good scientific understanding of the behavior of the exploited stocks is prioritary in order to implement management strategies. Action plans should focus in maintaining sustainable fishery practices and targeting numerically robust stocks as a priority to implement global management plans where dolphinfish populations are included. For the Eastern tropical Pacific further research is needed in order to evaluate if the temporal stocks found in this study are migrating from neighboring countries, to prevent fishery depletion. Lack of knowledge and prevention produced the fishery decline of Atlantic Bluefin tuna in Norwegian and German fisheries, where the lack of migration of maturetunas was explained by possible overfishing on local areas [32]. This is important information to be considered since dolphinfish populations also exhibit migration patterns and are targeted in different fishing grounds in many other countries in the Pacific (Mexico, Panama, Ecuador) [14].
For practical purposes, the direct conservation of particular alleles, or their frequencies would be almost impossible. Broader criteria as exploitation from numerically robust stocks, would reduce the impact on stock demographic dynamics, allowing the continuity of inter-stock exchange of genes and promoting this way the maintenance of high levels of abundance in all stocks [33]. In the specific case of dolphinfish, multiple fisheries compete to catch fish in different countries. Considering dolphinfish are moving in the Pacific and each country has not only different management strategies but a different total allowable catch (TAC), the combined numbers of TAC would end reducing the entire common-pool fishery and threatening the stock size [1]. Management authorities should seek a combined effort to reach sustainable fisheries
Although at this time we have no evidence to define if individuals collected in distant months of the year belong to reproductively isolated units, we suggest assigning the term "temporally defined stocks" to the population substructuring detected in this study. This information should be then considered before establishing a management plan for this fishery to ensure its long-term sustainability. Further research with a broader sampling and possible tagging methods might give a better understanding of the possible movement pattern of dolphinfish stocks in the Pacific.
To Know More About Journal of Oceanography Please Click on: https://juniperpublishers.com/ofoaj/index.php
To Know More About Open Access Journals Publishers Please Click on: Juniper Publishers
Comments
Post a Comment